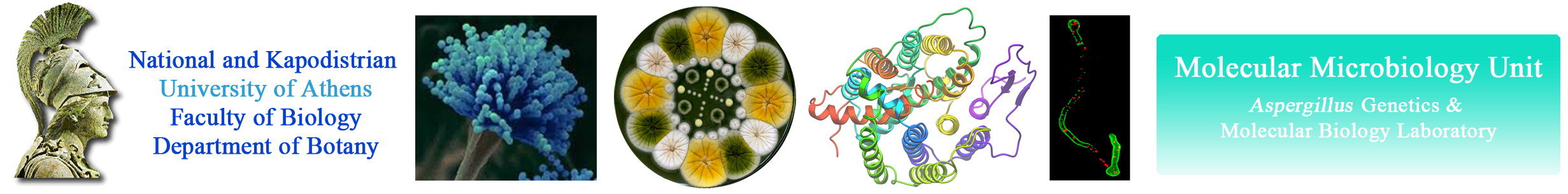
Why Aspergillus as a Model System
In 1953 Pontecorvo published the compendium of the genetics of the ascomycete fungus Aspergillus nidulans (1). Since 1984, after the development of transformation protocols, A. nidulans has also become a model microbial system for molecular biology and reverse genetics approach. Practically, most of what can be done in Saccharomyces cerevisiae, can be done in A. nidulans. What clearly distinguishes A. nidulans is its mode of polar life. Cell polarity is, for example, highly demanding in special mechanisms for rapid protein exocytosis and endocytosis along very long cells, the apical parts of which possessing additional needs on membrane trafficking, as compared to sub-apical parts or non-polar cells like yeasts. In fact, genetic evidence in filamentous fungi shows that rapid endocytosis of membrane proteins followed by their re-delivery to the plasma membrane generates and maintains polarity. Interestingly, the molecular mechanisms underlying fungal polarity are analogous or/and homolous to those found in the cells of higher animals, such as mammalian neurons.The beautiful and sophisticated system of A. nidulans genetics, molecular and cell biology led to the analysis of multiple metabolic and cellular processes. It suffices to highlight the identification by Claudio Scazzocchio, Herb Arst and co-workers of the crucial actors of nitrogen metabolite and carbon catabolite repression (2), the work of Ron Morris and his school (3), which matches the Nobel winning work of Paul Nurse and Lee Hartwell, the dissection of conidial development initiated by Bill Timberlake with the introduction of the methodology of cascade hybridisation (4), and the recent work of Miguel Angel Penalva on membrane trafficking, Golgi dynamics and endocytosis (5). Other Aspergilli are used in biotechnological processes that range from the production of soja sauce and sake (A. soyae, A. oryzae), to the production of citric acid (A. niger) and the production of heterologous proteins. A. fumigatus has graduated, thanks to immunosuppression, from rare pathogen to one of the most common causes of fatal nosocomial infection. A. flavus, a noxious contaminant of food-stuffs, produces aflatoxin, the most powerful carcinogen known. A. sydowi is an insidious pathogen of gorgonian corals. A recent issue of Nature carries three articles describing the genomic sequences of A. nidulans (6), A. oryzae (7) and A. fumigatus (8). The differences between A. nidulans and the other two Aspergilli match the differences found between fish and humans, which are separated by 500 million years, suggesting a very rapid rate of divergence in this genus. The A. oryzae genome has 12074 predicted proteins as compared with 9926 for A. fumigatus and 9541 for A. nidulans. The excess of A. oryzae putative genes concerns mainly secondary metabolism, and may be due to horizontal gene transfer, including from prokaryotes. A. nidulans is homothallic, but there are heterothallic species of Aspergillus, while no apparent sexual cycle has been described for A. fumigatus or A. oryzae (9). Particular for the interests of our lab, which mostly concern genes and proteins involved in purine or amino acid transport and metabolism, A. nidulans provides unique advantages related to growth phenotypes, already available strains, specific and direct genetic screens and the development of easy and fast metabolite uptake studies.
References
-
(1) Pontecorvo G., Roper J. A., Hemmons, L. M.., MacDonald K. D. and Bufton A. W. J. (1953) The Genetics of Aspergillus nidulans. Adv. Genet. 5, 144-238
-
(2) Arst H. N., Jr. and Cove, D. J. (1973) Nitrogen Metbolite Repression in Aspergillus nidulans. Mol. Gen. Genet. 126, 111-141
-
(3) Morris, N. R. and Enos, A. P. (1992) Mitotic gold in a mold. Trends Genet. 8, 32-37
-
(4) Timberlake, W. E. (1990) Molecular Genetics of Aspergillus Development. Ann. Rev. Genet. 24, 5-36
-
(5) Pantazopoulou A, Penalva MA. (2009) Organization and dynamics of the A. nidulans Golgi during apical extension and mitosis. Mol Biol Cell. 20: 4335-7
-
(6) Galagan J. E. et al. (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1106-1115
-
(7) Machida, M. et al. (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157-11661
-
(8) Nierman, W. C. et al. (2005) Genomic sequence of the pathogenic and allergenic fungus Aspergillus fumigatus. Nature 438, 1151-1156
-
(9) Geiser, D. M., Timberlake, W. E. and Arnold, M. L. (1996) Loss of Meiosis in Aspergillus. Mol. Biol. Evol. 13, 809-817
Current Interests of the Lab
Our group is primarily interested in several aspects concerning the expression, function, cell biology and evolution of transport proteins. Our model organism of choice is the non-pathogenic, filamentous fungus Aspergillus nidulans, a classic model genetic system, since the 1950's. Since the 90's, several A. nidulans transporters specific for purine, pyrimidine, nucleoside or amino acid transport have been cloned and studied in respect to their transcriptional, post-translational and cellular control of expression. The principal current interest of the lab is to use A. nidulans as a model system for:
Genetically and biochemically dissecting the structure-function relationships underlying purine transporter topogenesis, function and specificity
Identifying the pathways and molecular mechanisms involved in the membrane trafficking and endocytosis of specific transporters in response to various physiological, developmental and genetic signals.
Structure-function relationships in purine and amino acid transporters
-
Which residues make contact with the pyrimidine moiety of purines (we already know the residues making contact with the imidazol moiety of purines)?
-
Which residues are dynamically involved in substrate translocation?
-
How can we further convert the specificity of UapA, in order to mimic its mammalian homologues (SVCT-1 & -2) transporting vitamin C (ascorbic acid)?
-
Which residues/motifs are necessary for ER-exit, Golgi-exit and trafficking to the plasma membrane (see also below)?
-
Which residues or domains are involved in UapA oligomerization and what is the role of oligomerization in the function or trafficking of the transporter (see also below)?
-
In collaboration with the group of Ass. Prof. B. Byrne (Imperial College, London), we develop protocols for high-level expression/purification of UapA and bacterial homologues for downstream crystallographic or NMR studies.
-
In collaboration with the group of Prof. E. Mikros (School of Pharmacy, University of Athens), we are continuing mutational studies and molecular modeling in: i) the purine-pyrimidine-allantoin transporter family (NCS1), including several A. nidulans members cloned and studied in our lab (FurD, FurA, FcyB), ii) the Nucleobases-Ascorbate Transporter (NAT/NCS1) family. For both these families, general 3D templates have become recently available through crystallographic studies on homologous prokaryotic permeases. For all the above goals we are openly collaborating with Prof. S. Frillingos (Medical School, University of Ioannina), who is interested in bacterial members of the NAT/NCS1family.
Membrane trafficking and endocytosis of transporters
-
What is the role of lipid composition in transporter trafficking, endocytosis and vacuolar degradation?
-
How mutations resulting in transporter intrinsic instability lead to enhanced endocytosis and vacuolar degradation?
-
Which cargo cis-acting elements are involved in the "molecular conversation" with the trafficking machinery?
-
Which are the pathways followed, after exit from the Golgi, of a newly made transporter under different physiological conditions or in specific mutants?
-
What type of ubiquitination (mono-, multiple-, poly-, K63 chains) is involved and at which cellular compartments this takes place during transporter trafficking, in response to different physiological conditions or in mutants affected in trafficking?
-
Do different transporters follow similar pathways or use similar mechanisms for trafficking? Most of our initial studies employ as a model cargo the uric acid permease UapA. Using the originally obtained knowledge, subsequent studies will investigate the trafficking of several other transporters (UapC, FurD, AzgA, FcyB, CntA, PrnB, AgtA) readily available in our lab.
Transporters and channels play crucial roles in many cellular and physiological processes, through their action as mediators of cell communication with the extracellular environment. Despite their importance, the mechanism of transport function and specificity is very little known, due to the difficulties in obtaining X-ray diffraction-quality crystals and in applying NMR methods for the study of polytopic membrane proteins. Furthermore, the few currently known structures come exclusively from bacteria/archea, and in the majority of cases are not associated with functional studies. An alternative approach to understand structure-function relationships of transporters and channels is through functional analysis of mutated or molecularly modified versions of these proteins in easily manipulated systems. The magnificent work of R. Kaback and his group with the LacY lactose permease of Escherichia coli constitutes a unique paradigm in biology. In this case, solely through the analysis of mutations, biochemical and biophysical approaches and the development of Cys-scanning mutagenesis, a model was proposed on how a transporter works, even before the eventual crystallization of LacY (Guan L, Kaback HR. 2006. Annu Rev Biophys Biomol Struct, 35:67-91).
The contribution of our lab in this direction concerns the use of mostly classical and reverse genetics and kinetic modelling to understand how mutants of the UapA uric acid-xanthine transporter (for reviews see Gournas et al, 2008, Mol BioSyst, 4: 404-416; Diallinas & Gournas 2008, Channels, 2, 363-372) or the PrnB proline transporter work (Vangelatos et al. 2009. Mol Membr Biol 26:356-70 ). In the case of UapA, our favourite molecule, we have proposed models on how the transporter recognizes and translocates its substrates, and presented data on which amino residues are involved in substrate selection, binding and transport, which amino acid residues are key elements for protein stability and trafficking to the plasma membrane, how we can engineer new UapA versions transporting potential antifungal drugs and how domains external to the substrate biding site act as filters or gates affecting UapA substrate specificity.
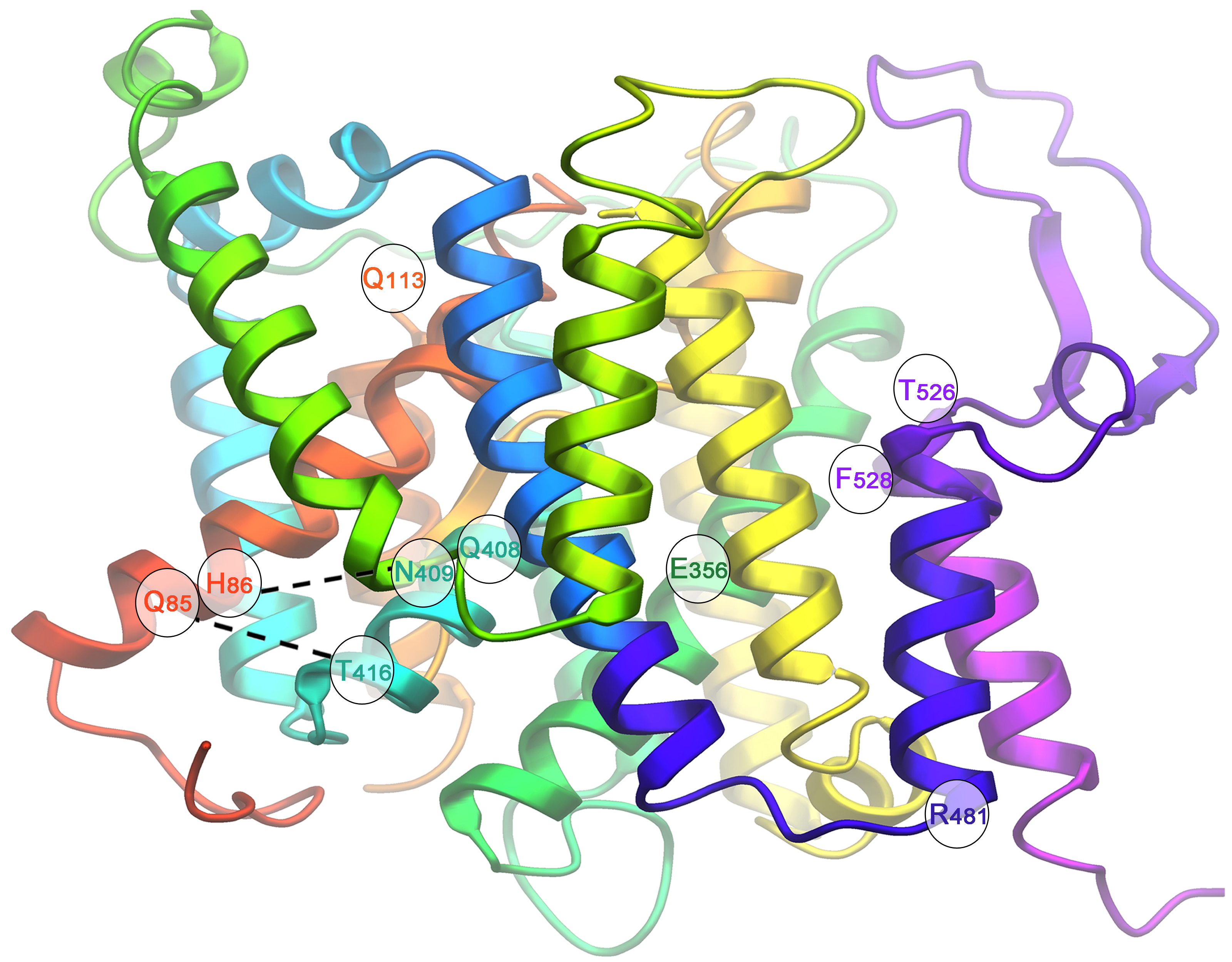
Through our work, UapA has become the prototype molecular model for understanding structure-function relationships in Nucleobase-Ascorbate Transporters (NAT/NCS1), which constitute an important and ubiquitous transporter family of unknown 3D structure (for reviews and more recent work see: Gournas et al, 2008, Mol BioSys, 4: 404-416; Diallinas &. Gournas 2008, Channels, 2, 363-372, Kosti et al. 2010. J Mol Biol. 397:1132-43. See also list of publications).
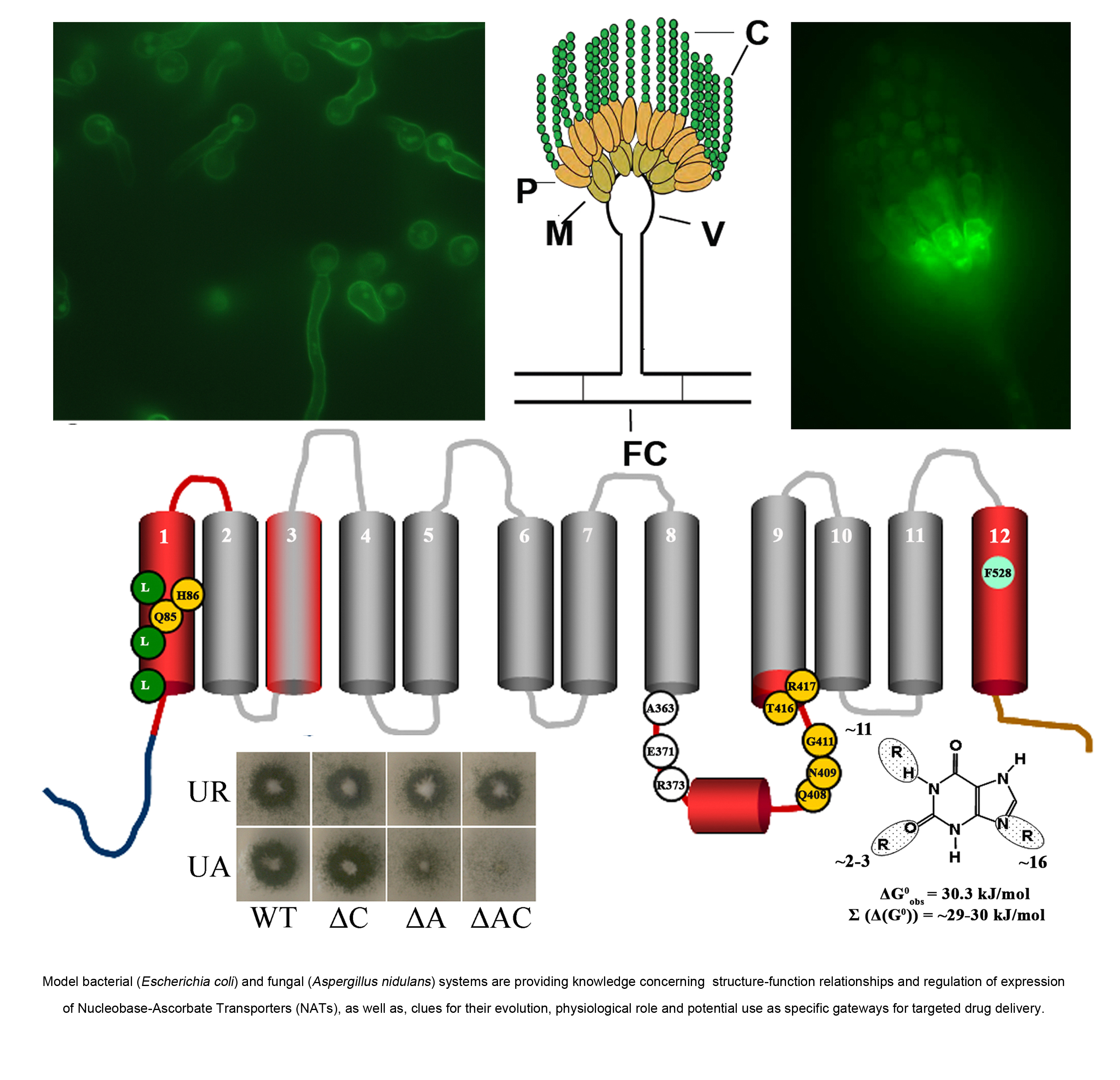 We are currently asking the following questions:
We are currently asking the following questions:
Eukaryotic polytopic membrane proteins, such as transporters, channels or receptors, are co-translationally inserted into the ER membrane through the action of the so-called translocase complex. This mechanistic step seems to be driven by the energy of polypeptide synthesis in the ribosome and several sequence-independent, amphipathicity-dependent, cis-acting elements on the protein cargoes. Once a membrane protein is properly folded within the ER membrane, an important check-point by itself, it then follows a vesicular or tubular trafficking pathway, initially towards the Golgi, and subsequently towards the endosomal pathway, the vacuole or the plasma membrane. This long sub-cellular journey (exocytosis) is dynamically controlled in response to multiple and overlapping developmental and physiological signals, rather than being a default process. Such signals not only promote or arrest exocytosis of a protein towards its target membrane, but can also promote its rapid endocytosis from the plasma membrane, which can lead to degradation in the vacuole or recycling back to the cell surface. The sum of complex processes underlying membrane protein exocytosis and endocytosis is called membrane protein trafficking. In this process, both cis-acting elements on the cargo proteins and trans-acting factors need to be orchestrated in a sequential and flexible manner to achieve proper trafficking. Moreover, it has been recently shown that the lipid composition of membranes, which by itself is dynamically controlled in response to various signals, plays a pivotal role in protein trafficking. The dynamic control of the trafficking of proteins such as transporters, channels and receptors, proteins, which play essential roles in the uptake or efflux of metabolites and drugs or the transmission of molecular signals, constitutes a rapid, flexible and very efficient regulatory mechanism critical for cell homeostasis and for the communication of cells with their environment.
Transporters and channels, but also other membrane proteins, are very often subject to tight regulation allowing cells to adapt to a changing environment. This is particularly well illustrated in the yeast S. cerevisiae in which several plasma membrane transporters have been studied. The intracellular trafficking of these proteins is typically controlled according to substrate availability, general nutrient supply conditions and/or stresses. Strikingly, the mechanisms controlling transporter trafficking are essentially conserved from fungi to mammals. In fact, studies on the intracellular trafficking of yeast permeases have contributed to revealing the central role played by the small protein ubiquitin (Ub), a sorting signal of eukaryotic membrane proteins. In yeasts and A. nidulans the covalent attachment of Ub on cargoes depends on Rsp5/HulA, a Ub ligase of the Nedd4 HECT family. Recent studies suggested a general model in which different Rsp5/HulA adaptor proteins recognize different transporters, or the same transporter in response to different stimuli. Finally, llipid rafts, formed by the lateral association of sphingolipids and cholesterol (mammals) or ergosterol (fungi) in the external membrane leaflet have been implicated in transporter traffic and cell signalling in mammalian cells and yeast (for further reading see several excellent articles and recent reviews from the labs of R. Hagauenauer-Tsapis, B. Andre, C. Kaiser, H. Pelham and S.D. Emr).
Several genetic and molecular tools for specific sub-cellular organelles/compartments have also been developed for A. nidulans (mostly in the lab of M.A. Penalva) and several transporters of purines, pyrimidines and amino acids, belonging to evolutionary discrete families, have been used as protein cargoes to study endocytosis in response to a shift in nitrogen source or excess substrate (our lab). As a consequence, important aspects of trafficking mechanisms have been revealed in this model fungus. The primary contribution of our lab in this direction is the identification of two distinct mechanisms controlling transporter down-regulation by endocytic internalization. The first occurs in response to a shift from poor to rich nitrogen media (ammonium ions) and the second in response to substrate excess (Valdez-Taubas et al. 2000, Fungal Genet Biol. 30:105-13; Pantazopoulou et al. 2007, Fungal Genet Biol. 44: 627-40; Gournas et al. 2010, Mol Microbiol. 75: 246-60). In the case of the UapA transporter, both mechanisms are dependent on HulA-dependent ubiquitination of a single Lys residue (K572) at the C-tail of the transporter. However, substrate-induced endocytosis, unlike ammonium-induced internalization, takes place only for active molecules. The use of specific functional mutations of UapA has shown that protein movements associated with the transport process constitute the upstream signal for substrate-induced endocytosis. Interestingly, active transporter molecules can elicit the endocytosis of inactive molecules in trans, suggesting that transporters oligomerize or are organized in membrane microdomains.
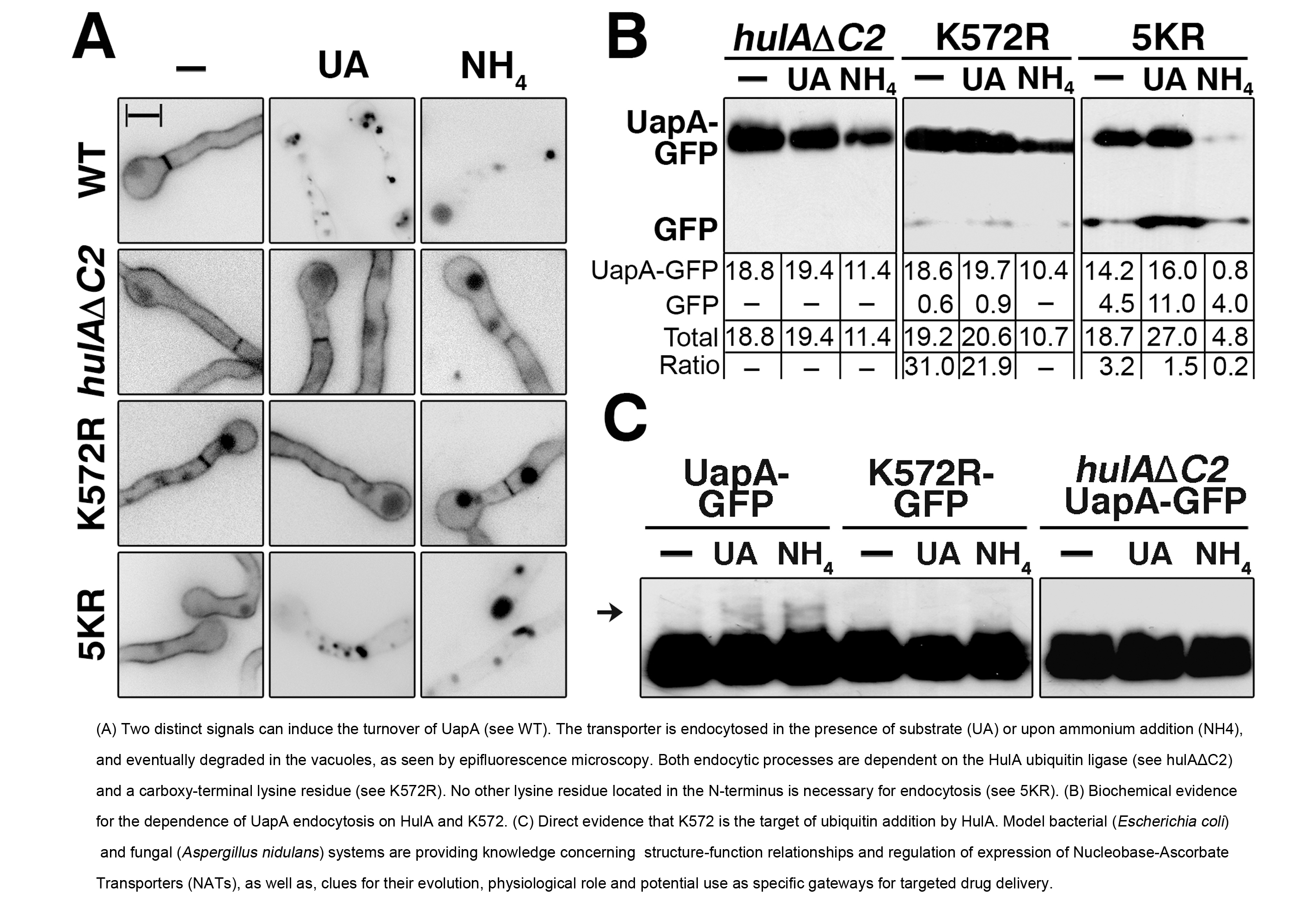 Some of our current specific questions are the following:
Some of our current specific questions are the following: